Introduction to Protein Modeling Projects
Welcome to the Protein Modeling Projects Page at NSUWorks. NSU Halmos College of Arts and Sciences faculty members, Dr. Arthur Sikora, Department of Chemistry, Engineering and Physics, and Dr. Emily F. Schmitt Lavin, Department of Biological Sciences, created this site as a repository for research projects in the field of protein modeling.
This site documents research primarily conducted by students enrolled in the honors course, HONR 1010G: Introduction to Protein Modeling which began in Fall 2021. Sikora and Schmitt were inspired by
3D Molecular Designs.
Projects prior to the course, are linked here as abstracts from the Annual Meeting of the Association for Biochemistry and Molecular Biology: 2017, 2018a, 2018b, 2019, 2020, and 2021. Projects in this series include the presentation of results from teams of student researchers investigating protein modeling tools to describe their molecular “stories” of interest. We invite you to enjoy reading the results of research into these molecular stories told through protein models.
-

Modeling the Extracellular Matrix-Like Retinal Protein Pikachurin and its Binding to α-Dystroglycan and GPCR (GPR179)
Mohit Belur, Khushi Raval, Riya Chelagiri, Rani Mehta, Emily Schmitt Lavin, and Arthur Sikora
2025
-

Modeling the Disruption of Binding of Three IgE Antibodies with Six Mutated Interaction Sites on the Arah2 Allergen
Hansi Parmar, Aashi Chhabra, Jyothi Vivekananda, Amaan Khan, Arthur Sikora, and Emily Schmitt Lavin
2025
-
Modeling OCT3-Mediated Buprenorphine Transport as a Mechanism for Salivary Accumulation and Oral Toxicity
Veda Pesala, Laasya Vemparala, Mehtab Singh, Sydni Gunpath, Emily Schmitt Lavin, and Arthur Sikora
2025
-

Modeling the Binding of Salinosporamide A (MZB) to the Human 20S Proteasome for Potential Cancer Treatment
Jayden J. Ross, Giezel Medina, Audrey Anand, Jayant Ari, Emily Schmitt, and Arthur Sikora
2025
-

Molecular Consequences of PCOS-Linked Aromatase Variants: Structural Modeling and Inhibitor Docking
Kiran S. Sajnani, Bharath Kumar Reddy Burri, Yuktha Milukuri, Preya Patel, Marqus Colón, Emily Schmitt Lavin Ph.D., and Arthur Sikora Ph.D.
2025
-

Modeling the Binding of Five Selective Serotonin Reuptake Inhibitors (SSRIs) to the Human Serotonin Transporter (hSERT) to Allosteric and Central Binding Sites
Alessandra Alvarez Coke, Ivanna Mathew, Srika Saai Talanki, Emily F. Schmitt Lavin, and Arthur Sikora
2024
-
Modeling the Function of Ryanodine Receptor Type 2 and its Role in Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT) Heart Disease
Harani Ritesh Athwale, Dillen-Preetesh Jayanti Patel, and Sai Puneeth Kothuru
2024
-
Modeling Isocitrate Dehydrogenase 2 Mutations in Acute Myeloid Leukemia: Exploring how the drug AG-221 Inhibits 2-hydroxyglutarate Production and Restores Blood Cell Differentiation
Bhumika Balani, Jay Matta, Nitya Punjal, and Brianna Bourke
2024
-

Modeling Nicotine, AT-1001, and Varenicline at the α3β4 Nicotine Acetylcholine Receptor
Japji Madan, Andersen Cheong, and Mourya Khandai
2024
-

Modeling the Binding of Dendrin and PTPN14 to KIBRA
Aniruddh Mehra, Anna Maria Weber, and Chandralekha Sai Mekala
2024
-
Using 3D Modeling to Describe the Electromotility of the Outer Hair Cell Protein Prestin, and its Role in Sound Perception Among Mammals
Marqus Colon, Syed Hussain, Russel Robert Reside, Emily Schmitt-Lavin, and Arthur Sikora
2023
-
Modeling the Binding of Tetrodotoxin and Saxitoxin to the Nav1.7 Voltage-Gated Sodium Channel
Isabella G. Fiore, Smrithi Mukund, and Laasya Buddharaju
2023
-
Modeling Cysteinyl Leukotriene Receptor Antagonist KNW for Possible Optimized Asthma Treatment
Shalet James, Sreejani Jonnalagadda, Emily Schmitt Lavin, and Arthur Sikora
2023
-
Modeling Calcium Binding to Yersinia pestis Type III Secretion Needle
Kaya Olszewski, Priyanka Prasanna, Chetana Movva, Arthur Sikora, Emily Schmitt Lavin, and Julie Torruellas Garcia
2023
-
Identifying the Binding Residues on CYP3A4 to Naringin using Protein Modeling and Docking
Parth Shah, Vraj Patel, Sanjana Ananthula, Emily Schmitt Lavin, and Arthur Sikora
2023
-
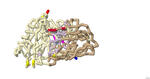
Binding of Beta-site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitor Aminoquinoline (68K) for Possible Treatment of Alzheimer's Disease
Juhi Dalal, Shreya Averineni, Pranav Madadi, Emily S. Lavin, and Arthur Sikora
2022
-
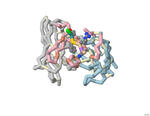
Comparing Effectiveness of Two Antibodies (Aducanumab and Gantenerumab) on Reducing Amyloid-Beta Plaques
Nikhila Paleati, Pranav R. Neravetla, Akhil B. Godbole, Emily S. Lavin, and Arthur K. Sikora
2022
-
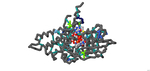
Investigating The Mechanisms of Active Site Mutations to the 1T9G WT MCAD Protein to Better Understand Medium Chain Acyl-CoA Dehydrogenase Deficiency (MCADD)
Omar E. Saleh, Rhea Khatiwala, and Jeremy Ignatius
2022
-
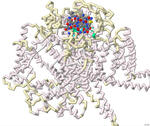
Modeling the binding of ω-conotoxin and other toxins to the N-type voltage-gated calcium channel
Serena Sha, Sophie Welch, and Ashley Guillen-Tapia
2022
-
Exploring structural differences between antagonistic peptides for the development of orally bioavailable PCSK9 inhibitors
Bhavya Soni and Pritika Vemulapalli
2022
-

How can we design an inhibitor with an enhanced binding affinity that is selective for MMP12 ?
Aisha Y. Abdool, Lyla Abbas, Tassnime Sebaei, Emily Schmitt, and Arthur Sikora
2021
-

Comparing the Effectiveness of Ivacaftor and GLPG1837
Jordan Nichole Carreras, Saimi Reyes, Vibha Sankavaram, Emily Schmitt Lavin, and Arthur Sikora
2021
-
Limiting Iron Acquisition of E. Coli With Anti-TonB1 and AntiTonB2
Jose Diaz, Ryan Luib, and Seethal Doki
2021
-
Highlighting How the Structure of Marine Bacterial Laminarinase Can Improve Biogeochemical Cycling During Global Climate Change
Heidi Hellenbrand, Rachel Harris, and Chino Villanueva
2021
-
Investigating the Structure of Potential New Drug to Treat Sickle Cell Anemia through Inhibition of the Polymerization of Hemoglobin S
Brianna M. Lacasse, Isadora R. De Abreu, and Rathika Manikandan
2021


