Files
Submission Date
Fall 2022
Abstract
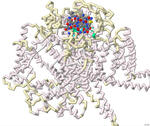
Approximately 1.5 billion people in the world suffer from chronic pain, persistent pain that carries on for longer than 12 weeks despite medication or treatment. Management of chronic pain typically includes the use of non-steroidal anti-inflammatory drugs (NSAIDs) or prescription pain medications, including opioids. An alternative therapy derived from conotoxins, toxins released from marine predatory snails in the family Conidae, was approved for the treatment of severe chronic pain in 2004. This pharmaceutical has the trade name Prialt and is also known as ziconotide. Once ziconotide is in the human body, it acts as a channel blocker of the N-type voltage-gated calcium channels, also known as Cav2.2. The literature and related PDB files were manipulated using PyMol and Jmol to create a 3D-printed model to explain the molecular story behind how a particular conotoxin binds to a calcium-gated ion channel. Additionally, computer visualization tools were used to show how several related toxins from other organisms would be expected to dock to the calcium ion channel. The 3D-printed model highlights specific features that contribute to the ω-conotoxin (MVIIA) binding to the calcium channel alpha 1B subunit as described in the literature (PDB: 7MIX). These conotoxins have a very characteristic disulfide bond linkage pattern which plays a role in the correct folding of the peptide and stabilization of its structure. In MVIIA, the non-cysteine amino acids form unstructured loops affecting binding affinity and calcium channel-blocking activity. Of particular interest is the second loop located between Cys8 and Cys-15. It appears to be exceptionally important in directing selectivity toward N-type calcium channels and away from P/Q-type calcium channels. Ziconotide does not directly seal the entrance to the vestibule of the selectivity filter, but it blocks ion entrance by neutralizing the outer electronegativity and sterically hindering the ion access path to the entrance of the selectivity filter. Salt bridges are formed between Arg10 and Tyr13 on ziconotide and Asp664 of the channel. Four of the eight ziconotide-coordinating residues, Thr643, Asp1345, Lys1372, and Asp1629 in Cav2.2 are not conserved in other calcium channels which may explain the subtype specificity of pore blockage by ziconotide. The EEEE motif consisting of Glu314, Glu663, Glu1365, and Glu1655, determines the Ca2+ selectivity. Also included in the model are the receptor's alpha helices and bound calcium ion. The N terminus and C terminus of the receptor are labeled in blue and red respectively to orient the model.
No crystal structures are available for ω-conotoxins bound to several other types of N-type calcium channels. To investigate the potential calcium channel blocking properties of conotoxins MVIIC, GVIA, MoVIB (from the cone snail, Conus magus) and ω-agatoxin IVA (from the spider, Agelenopsis aperta), the computer-based tool ROSIE was used to simulate binding of these peptides to the Cav2.2 channel. As expected, the toxin shown in the crystal structure of 7MIX, bound best to the Cav2.2 channel. The toxins MVIIC, GVIA, and MoVIB bound with lower affinity. The agatoxin IVA did not have any relevant binding to this calcium channel. Overall, protein modeling allowed for a deeper understanding of how conotoxins bind to and block the calcium channel possibly leading to additional therapeutic approaches to pain relief.
Support or Funding Information:
This work was made possible by funding through the National Science Foundation, Division of Undergraduate Education (NSF-DUE) grant number 1725940 for the CREST Project. Nova Southeastern University’s Farquhar Honors College and Dept. of Biological Sciences also provided support. Protein model printing was made possible by 3d Molecular Designs.
Recommended Citation
Sha, Serena; Welch, Sophie; and Guillen-Tapia, Ashley, "Modeling the binding of ω-conotoxin and other toxins to the N-type voltage-gated calcium channel" (2022). Protein Modeling Reports. 9.
https://nsuworks.nova.edu/protein_modeling_reports/9